Services
– Agilent Bioanalyzer
• DNA-High Sensitivity Kit
• Small RNA kit
• RNA 6000 Pico kit
• Online Submission Request
• Physical Submission of Samples
• Data Analysis
• Other Bioanalyzer Kits
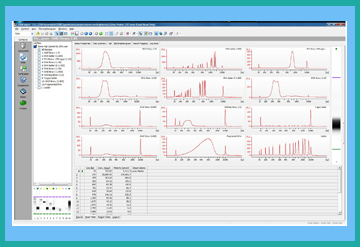 Enlarge image of DNA High Sensitivity results.
Enlarge image of DNA High Sensitivity results.
DNA-High Sensitivity Kit
This kit is kept in stock at the Genomics Core; other types of kits (see Other Bioanalyzer Kits) can be purchased from Agilent.
Visit the Agilent website for product specifications of its
DNA and RNA kits; highlights for the DNA HS kit are described below.
Chip capacity: 11 “sample” wells. However, unless you are mostly looking at the
fragment size distribution and the exact DNA concentration is not critical, it's best to process
each actual sample at least in duplicate, if not triplicate. This is due to the small volumes
being pipetted, for which even tiny offsets from the intended volume can lead to fairly large
accuracy deviations. If replicates are done, the number of samples/chip will be correspondingly fewer.
Tubes: Samples must be submitted in 0.2-ml tubes:
– Separate tubes with hinges are preferred.
– Tubes with narrow bottoms are preferred.
– Do not use low-profile tubes.
Sample volume: 1 µl of sample is loaded in each well on a chip.
- Preferred Option: Provide exactly 1-µl of each sample (or replicate sample) per tube, as discussed in Loading Agilent Bioanalyzer chips... Genomics Core style!. Note: With the Preferred Option, duplicates are simply a second tube containing another µl of the same sample.
- Discouraged Option: This option is allowed, but definitely discouraged, as it involves more effort and chances for error. For each sample, provide 2-3 extra microliters to allow for accurate pipetting of a single µl. For running
singlets, that’s 3-4 µl total; for duplicates, it would be 5-6 µl (and so on).
Sample Concentration: In the context of ladder peaks (which are ~10-15 bp wide), DNA samples can range from 5-500 pg/µl. However, the minimum and maximum values are higher for smears as the total fluorescence is spread over a wider zone.
For a well-defined DNA smear (e.g., ~50-75 bp wide), a decent reading often can be achieved at concentrations as low as ~100 pM. For broad smears, 5 ng/µl is likely the absolute maximum rational concentration and values of ~1-3 ng/µl will likely be more suitable; see the following two images for examples with broad smears.
Bioanalyzer_Concentration_Guidelines_1.jpg; Bioanalyzer_Concentration_Guidelines_2.jpg
If you don’t already have a good idea of the DNA sample concentrations, it can be beneficial to determine if dilutions are needed by estimating sample concentrations with the Genomics Core's Qubit dsDNA kit (or, even a Nanodrop... as long as you pay attention to the caveats noted at Nanodrop Nucleic Acid Guide.pdf). You might also consider running serial dilutions on the chip to ensure that at least one well is within the right range.
Buffer: There should be at least some salt in the buffer; the maximum salt is 10 mM Tris
and 1 mM EDTA. Further, all DNA samples loaded onto the same chip should be in the same buffer;
substantial differences in buffer compositions among samples can result in chip failures.
Beads & Particulates: Residual beads from a DNA-cleanup step or even particulates from a column can wreak havoc on a Bioanalyzer chip. It is highly recommended that samples be centrifuged and then (if magnetic beads were used) placed on a magnet for 2 minutes prior to taking the 1-ul aliquot (from just below the liquid surface) for the Bioanalyzer assay.
Fragment size range: ~50-7000 bp (size markers are
at 35 & 10,000 bp); however, from a practical standpoint, you should aim for a maximum fragment size of ≤2000-bp so that your DNA fragments are unlikely to elevate the baseline in the region of the Upper Marker.
Upper Marker: DNA samples concentrations are estimated based on the area under the Upper Marker; if the baseline is elevated in that region, it can adversely affect concentration estimates. Further, substantial elevations in the baseline under the Upper Marker can adversely affect results in even neighboring wells on the chip. For amplified libraries, over-amplification is probably the most common reason that non-baseline readings occur in the Upper Marker region (aside from the Upper Marker itself). In the case of sheared genomic DNA, under-shearing of the sample can be the problem.
↵ Back to Top
Small RNA kit
The Small RNA kit is recommended for analysis of small/micro RNA molecules (e.g., separation of miRNA smears from tRNA). As we do not stock the Small RNA kit, you will need to coordinate with the Core at least 2-3 weeks ahead of when your small RNA samples would be ready for analysis.
↵ Back to Top
RNA 6000 Pico kit
This kit is used for mRNA, Plant RNA, and Eukaryote or Prokaryote Total RNA.
Chip capacity: 11 “sample” wells.
Submission Lead Time:
– If you expect to run a large number of chips (e.g., ~5 chips... or ~55 samples) at one time, please notify the Core at least two weeks ahead to allow for ordering a new kit should stock be low.
– Preparing to run an RNA Bioanalyzer chip takes >1-2 hours; thus, samples are more likely to be run the same day if submitted early in the day (i.e., before noon – by 1:00 p.m., at the latest).
– If next-day service is okay, please state that in your Client Memo of the online submission; further, state whether you would prefer that the samples be refrigerated or frozen until run.
Tubes: Samples must be submitted in 0.2-ml tubes:
– Separate tubes with hinges are preferred.
– Tubes with narrow bottoms are preferred.
– Do not use low-profile tubes.
Sample volume: 1 µl of sample is loaded in each well on a chip.
- Preferred Option: Provide exactly 1-µl of each sample (or replicate sample) per tube, as discussed in Loading Agilent Bioanalyzer chips... Genomics Core style!. Note: With the Preferred Option, duplicates are simply a second tube containing another µl of the same sample.
- Discouraged Option: This option is allowed, but definitely discouraged, as it involves more effort and chances for error. For each sample, provide 2-3 extra microliters to allow for accurate pipetting of a single µl. For running
singlets, that’s 3-4 µl total; for duplicates, it would be 5-6 µl (and so on).
Sample Concentrations: differ from the DNA HS kit — visit the Agilent website for
RNA 6000 Pico kit for complete product specifications or see Quick View Specifications (jpg).
Sample Type: It is essential to specify the type of RNA (mRNA, total RNA) being analyzed as well as the source (e.g., plant, eukaryote, prokaryote, yeast); please contact the Genomics Core for details.
Sample Buffer: The RNA Pico 6000 kit is extremely sensitive to salt concentrations; see Quick View Specifications. Ideally, samples should be eluted in either nuclease-free water or The RNA Storage Solution (ThermoFisher AM7000, 1 mM sodium citrate, pH 6.4).
Beads & Particulates: Residual beads from an RNA-cleanup step or even particulates from a column can wreak havoc on a Bioanalyzer chip. Prior to taking the 1-ul aliquot (from just below the liquid surface) for the Bioanalyzer assay, it is highly recommended that samples be centrifuged and then (if magnetic beads were used) placed on a magnet for 2 minutes.
RIN explained: The BMC article, The RIN: an RNA integrity number for assigning integrity values to RNA measurements, explains the RIN concept with respect to the Bioanalyzer assay; also, see below for various cautionary notes about RIN values and their usage. The RIN algorithm's partitioning of the RNA electropherogram is shown by this Quick View RIN Computation Graphic.
RIN or DV200: For Total RNA, the assays calculate RIN values by default; however, DV200 values can be calculated for some situations. If DV200 values are required, please specify that requirment in the Client Memo of your online submission.
RIN value exceptions: The Bioanalyzer RIN value algorithm was designed for mammalian tissues; thus, it does not necessarily give accurate estimates for a variety of other organisms (e.g., many insects, plants, prokaryotes) which may have very different total RNA profiles from mammals. For instance, while Drosophila total RNA does not have the ‘classic’ hidden break issue discussed below, the main peaks run differently from the typical mammalian RNA (see page 25, Bioanalyzer-TapeStation app-compendium. Various publications and online ‘chats’ about insect RNA generally say that you have to ‘eyeball’ the electropherograms, knowing what a good one looks like for your insect.
RIN — Cautionary statements by Agilent:
– The RIN can
: (1) assess RNA integrity; (2) compare RNA samples (e.g., before/after shipment, across samples from different labs); and (3) ensure repeatability of experiments (e.g., if specific RIN values are shown suitable for particular experiments, then that RIN can always be used for experiments using the same experimental design & tissues).
– The RIN cannot predict experimental success if no prior verification was done (
e.g., different RIN values might be required for microarray experiments vs. RT-PCR experiments or for 3'- vs. 5'-amplification).
– Validation: The computation of the RIN hasbeen validated only for
Eukaryote Total RNA Nano
samples. The 2100 Expert Software calculates the RIN for prokaryote and plant total RNA samples and for the RNA 6000 Pico assay – however, for these samples, the RIN has not been validated in extensive downstream experiments.
Sample Denaturation: The standard protocol calls for heat-denaturation of samples (70oC, 2 min) immediately prior to analysis. For a full discussion of the rationale for this step, see Stringent RNA quality control using the Agilent 2100 bioanalyzer (vendor pdf). In brief, the heat-treatment (HT) ensures elimination of secondary structures which might cause peaks to co-migrate or might prevent nicked RNA from separating during analysis. If separate RNA fragments co-migrate due to secondary structure, the RIN values will be incorrectly reduced; by contrast, if extraction of the RNA sample has caused nicking, the RIN values could be incorrectly high. Thus, the heat-denaturion step will be performed unless you specifically request its omission in the 'Client Memo' for your online Submission and put a note on the tape label for your samples.
There are two reasons to skip sample heat-denaturation:
– "Hidden Breaks": Many species (especially among protostomes – e.g., arthropods, molluscs, annelids) contain a "Hidden Break" in the large subunit molecule (23S, 25S, or 28S RNA). Heating the purified RNA may cause the large subunit to separate into two roughly equal-size fragments that co-migrate during electrophoresis with the small subunit molecule (16S or 18S RNA). If so, that will result in an erroneously low RIN value. For species that contain a "hidden break", omitting the heat denaturation step may result in a more normal RNA profile – it does, however, mean that RNA-extraction nicks might also be missed.
The list below provides some examples of non-protostome organisms known to have "hidden breaks"; clients are responsible for determining if their particular organism might have "hidden breaks":
Agrobacterium tumefaciens (Gram-negative soil bacterium).
Bombyx mori (silkworm moth).
Cyprinidae (fish, tetraploids).
Heterocephalus glaber (naked mole rat).
–
RNase contamination: If your samples contain RNase molecules (either from lack of purification or through contamination), the heat-denaturation step may cause further RNA degradation, resulting in a poor Bioanalyzer profile. By contrast, keeping the samples cold until loaded in the chip may give a better view of the RNA integrity following your extraction protocol. However, please note that such residual RNase activity will ultimately lead to poor downstream results... but, you will not be aware of the problem because you skipped the heat-denaturation step. So, there is a trade-off when omitting the heat-denaturation step.
Frankly, if the samples are free of RNAse contamination and are properly buffered, they should be perfectly fine being stored temporarily at room temperature. Thus, some sources recommend leaving your RNA samples at room temperature overnight before running them on the Bioanalyzer to verify that the RNA will remain intact for the downstream processing. An excellent buffer for this purpose is
The RNA Storage Solution (ThermoFisher, AM7000 – 1 mM sodium citrate, pH 6.5 ± 0.1). Although Agilent recommends resuspending RNA is pure nuclease-free water because RNA 6000 Pico kit is highly sensitive to salt concentrations, we have found that 'The RNA Storage Solution' has no adverse effects for this assay.
↵ Back to Top
Online Submission Request
Log In, go to "Request Form", & click on "Services & Loan Items":
- Make the necessary 'selections'.
- Quantity: refers to the 'chip' (not your 'samples').
- Client Memo (DNA & RNA), specify:
(1) number of samples (& names, if desired);
(2) "same-day service requested" or "next-business-day service okay"; and,
(3) whether samples should be refrigerated or frozen if they cannot be run the same day.
- Client Memo (RNA): The following information is required for selecting the correct Run protocol for your RNA samples. If the wrong protocol is used, your data may be compromised (see RNA 6000 Pico kit).
(1) Source of samples (e.g., prokaryotes, plants, eukaryotic "animals", insects) – some situations require detailed explanations (e.g., gut samples from eukaryotes might be dominated by prokaryotic RNA);
(2) 'mRNA' or 'total RNA'; and,
(3) whether or not samples should be heat-denatured prior to analysis (see 'Sample Denaturation' comments above).
- Click the 'check box', and submit your request.
If you make an error in your request, you may revise that request if you do so before we begin processing it.
You will receive an automated email when your samples are processed and your results are uploaded to the website (click on
'Submission History' and download the 'File').
Files will be “zipped”. Files will remain in your 'Submission History' for ≥90 days; if you need a copy of the data after it's purged, we can usually retrieve it from our archives.
↵ Back to Top
Physical Submission of Samples
0.2-ml Tubes: Individual or strip tubes are acceptable.
Label Submission: Upon successful submission of your online request, the 'Submission #" will appear on your screen. Then, label the 0.2-ml tube rack itself with:
- Submission #;
- Date;
- PI's Last Name;
- Submitter name or initials; and,
- Special processing requests (if any).
Delivery site: Transfer tubes to a Genomics Core rack (taking your own rack back to
your lab), and put samples in the Mini-fridge (by sink) in the Genomics Core (A628 Life Science Annex).
[Note: If the Core is locked, put your samples in the 'After-Hours' Mini-fridge located in the hallway (by Room A649).]
↵ Back to Top
Data Analysis
To examine and manipulate the resulting *.xad files, download the “Trial” version of the
Bioanalyzer software
(for Windows systems). Although this freeware cannot run the Bionanalyzer itself, it is fully functional for analyzing *.xad files.
The software can generate multiple types of output beyond what is shown above; further, samples can be overlain... even from
different chips (of the same assay type).
See Documents for Agilent Bioanalyzer for assistance.
↵ Back to Top
Other Bioanalyzer Kits
Click links to see examples of data (for a single sample) from some of the many assays
that are available with the Agilent Bioanalyzer.
- DNA_High-Sensitivity.jpg
- DNA_1000.jpg
- DNA_7500.jpg
- DNA_12000.jpg
- mRNA (nano-series).jpg
- mRNA (pico-series).jpg
- Small RNA (series II).jpg
- Plant RNA (nano).jpg
- Prokaryote Total RNA (pico).jpg
- Eukaryote Total RNA (nano).jpg
- Protein (High Sensitivity).jpg
- Protein (80 series).jpg
- Protein (230 series).jpg
- Cy5-labeled nucleic acids (nano.jpg)
↵ Back to Top


