Overview: LSU Genomics Core

Dr. Mark Batzer (1961-2026)
Mark was a devoted member of the LSU Science community for 25 years and had a profound impact on many across the College of Science, including countless students. One of the university’s most distinguished researchers with a total of 17 journal covers, he was internationally recognized for his work on transposable elements and their impact on eukaryotic genomes and genetic variation. Mark also founded the LSU Genomics Core in 2001. Further, through his generous support as Director and primary funding support of the Core, he subsidized the research of many other laboratories at LSU, contributing to numerous publications and the development of graduate students and post-doctoral staff throughout the Department of Biological Sciences and beyond. Mark will be deeply missed by all who had the privilege of knowing him.
Genomics Core Funding
Since 2001, under the auspices of Dr. Mark Batzer, the LSU Genomics Core has provided services to the LSU Baton Rouge campus under an operational model in which instrumentation, service contracts, and staff were provided without charge. In essence, clients reimbursed the Core solely for the actual costs of reagents or consumables used to provide their services. As shown by the GC 2021 Survey – Our Past & Future?, this model was well received; however, new funding sources are needed to sustain that level of support for research at LSU.
Visual Tour of Core
For a visual introduction to our offerings, view this Slide show (*.pdf) or this 90-second silent video. The video is best viewed in "full screen" mode; it is not downloadable.
Potential Instrument Acquisitions
Are you interested in having a new platform available "in your backyard"... the LSU Genomics Core? If so, please provide the information requested at our Potential Instrument Acquisitions page. Some examples of prospective instruments are listed there, with links to the vendor websites and brochures.
Who has access to the website?
 Open access is provided to protocols, FAQs, links, and most documents on our website.
The "Science Aid Center" provides extensive guidance, especially for DNA Sanger-Sequencing.
Experimental protocols, documents, software, and
informational sites are available through "Links". When practical, we will even respond to email requests by non-affiliated researchers for assistance with instruments or protocols.
Open access is provided to protocols, FAQs, links, and most documents on our website.
The "Science Aid Center" provides extensive guidance, especially for DNA Sanger-Sequencing.
Experimental protocols, documents, software, and
informational sites are available through "Links". When practical, we will even respond to email requests by non-affiliated researchers for assistance with instruments or protocols.
Who has access to the Core's services?
 Members of the College of Science (LSU—B.R.) are our primary clients; other local campus labs may have access if their Cores lack similar capabilities. Some equipment is available for use directly by researchers; other equipment is managed solely by Core staff — see listings below. People lacking a Genomics Core Login ID can view the online request options available to authorized users by visiting the "View Only" version of the Request Form Options page.
Members of the College of Science (LSU—B.R.) are our primary clients; other local campus labs may have access if their Cores lack similar capabilities. Some equipment is available for use directly by researchers; other equipment is managed solely by Core staff — see listings below. People lacking a Genomics Core Login ID can view the online request options available to authorized users by visiting the "View Only" version of the Request Form Options page.
How can I begin using the Core?
 New Login IDs can be obtained through Request a Login ID. However, if the lab's PI has never used the Genomics Core before, prior to issuance of the Login ID, you (and your PI) must first
meet with Scott to
discuss the Core’s regulations and how to access restricted portions of the Website.
Use of the 'self-serve' equipment requires an Advance Reservation, made through the
Biosci-Booked (Genomics) instrument reservation system. All other requests for supplies, services, or sequencing require submission of an online Request Form.
New Login IDs can be obtained through Request a Login ID. However, if the lab's PI has never used the Genomics Core before, prior to issuance of the Login ID, you (and your PI) must first
meet with Scott to
discuss the Core’s regulations and how to access restricted portions of the Website.
Use of the 'self-serve' equipment requires an Advance Reservation, made through the
Biosci-Booked (Genomics) instrument reservation system. All other requests for supplies, services, or sequencing require submission of an online Request Form.
regarding processing and completion of requests.
You must Request a Login ID if you:
Exceptions: Requests may be submitted under another person's Login ID under two circumstances:
1) Transient researchers who are visiting the laboratory for a limited time; and,
2) Undergraduates temporarily working under the specific direction of the person to whom the Login ID was issued.
Instruments, Services, & Links
Common Room (A624, Life Science Annex)
Next door to the Genomics Core (A628), there is a Common Room (A624) which houses additional equipment maintained by the Department of Biological Sciences. Please note that the Genomics Core has no information regarding Common Room equipment... aside from the following:
- LSA-624 Common Equipment (partial listing):
- Ice Machine (granular ice).
- Milli-Q Synergy UV (ultrapure, type 1 water).
- Thermo NanoDrop 2000 (micro-volume spectrophotometer).
- ChemiDoc XRS (photo documentation, uv-gels).
- Beckman centrifuges (high-speed centrifugation).
- Hood, standard (e.g., organic extractions).
- Jouan MSC 12 (biosafety cabinet).
ABI 3130xl Genetic Analyzers
 The 3130xl Genetic Analyzers (Applied Biosystems) can run a wide variety of sequencing
and fragment analysis applications. As of January 2020 (when security updates to Windows 7 ceased), we converted the computers for our instruments to Windows 10 (64-bit); for details on this process, please see my LinkedIn post, Converting an ABI 3130xl computer to Windows 10. Typically, our two 3130xl’s are set up for standard DNA Sanger-Sequencing (≤900 bp)
with POP7 polymer and 50-cm arrays, a combination which also performs well for fragment
analysis; upon special request, we can install 80-cm arrays (≥1000 bp reads) or
36-cm arrays (rapid sequencing and fragment analysis). Requests for sample processing and data retrieval are both handled online
through the Genomics Core website.
The 3130xl Genetic Analyzers (Applied Biosystems) can run a wide variety of sequencing
and fragment analysis applications. As of January 2020 (when security updates to Windows 7 ceased), we converted the computers for our instruments to Windows 10 (64-bit); for details on this process, please see my LinkedIn post, Converting an ABI 3130xl computer to Windows 10. Typically, our two 3130xl’s are set up for standard DNA Sanger-Sequencing (≤900 bp)
with POP7 polymer and 50-cm arrays, a combination which also performs well for fragment
analysis; upon special request, we can install 80-cm arrays (≥1000 bp reads) or
36-cm arrays (rapid sequencing and fragment analysis). Requests for sample processing and data retrieval are both handled online
through the Genomics Core website. — Electrophoresis-Only: Client performs the BigDye sequencing reaction, cleans the products, and resuspends them in (preferably) ABI Hi-Di formamide such that the samples are ready for electrophoresis on the 3130xl. Guidelines and requirements for DNA sequencing are outlined in Sequencing_Instructions.docx; additional assistance can be found in the Science Aid Center and Documents & Protocols (under the heading of "ABI 3130xl Genetic Analyzers").
— DNA Templates:
· Option #1 – Client submits cleaned-&-quantitated DNA templates (see Full-Service DNA Sequencing Instructions), with the Genomics Core performing the BigDye sequencing reactions and cleanup procedures, followed by sample electrophoresis on the 3130xl.
· Option #1 – Option #2: Alternatively, the DNA Templates can be cleaned by the Genomics Core prior to sequencing; see DNA Templates (at Service Levels: DNA Sequencing).
— Electrophoresis-Only: Samples must be submitted as completed reactions, including the addition of any necessary size standards.
— Partially prepared samples: Clients may submit raw PCR products (i.e., FA products) and request that the Core dilute/aliquot the FA products and add the size-standard to all wells... prior to performing electrophoresis on the ABI 3130xl.
— Sample Denaturation: Upon request, FA samples will be heated and snap-cooled immediately prior to loading them on the 3130xl.
— Sequencing or Fragment Analysis: All samples must be submitted in either strip-tubes (with detachable caps) or 96-well plates.
— DNA template purification: In some cases, users might request that the Genomics Core purify their DNA templates prior to sequencing. For processing with our EDTA-Ethanol precipitation protocol, samples must be submitted in either strip-tubes (with detachable caps) or 96-well plates. However, for processing with the Zymo DNA Clean & Concentrator-5 Kit, standard 0.5-ml or 1.5-ml tubes are preferred.
– GeneMapper Software v5
– SeqScape Software v3
– Variant Reporter v2
– Sequencing Analysis v6
↵ Back to List of Instruments & selected Services
Agilent Bioanalyzer
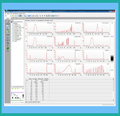 The Agilent 2100 Bioanalyzer system provides sizing, quantitation and quality control of DNA, RNA,
proteins and cells on a single platform, providing high quality digital data. The Genomics Core
currently stocks the DNA High Sensitivity kit; however, other kits can be ordered. The DNA-HS chip
provides critical Library QC data (quantitation AND fragment size distribution) for NGS experiments...
regardless of which NGS platform is used. For additional information on this platform and available
kits, see Bioanalyzer
options.
The Agilent 2100 Bioanalyzer system provides sizing, quantitation and quality control of DNA, RNA,
proteins and cells on a single platform, providing high quality digital data. The Genomics Core
currently stocks the DNA High Sensitivity kit; however, other kits can be ordered. The DNA-HS chip
provides critical Library QC data (quantitation AND fragment size distribution) for NGS experiments...
regardless of which NGS platform is used. For additional information on this platform and available
kits, see Bioanalyzer
options.
↵ Back to List of Instruments & selected Services
BluePippin (DNA Size Selection for NGS)
 The BluePippin.jpg has the capabilities of the Pippin Prep, plus pulsed-field electrophoresis for resolving and collecting high molecular weight DNA. Target sizes or ranges of sizes are entered in software, and fractions are collected in buffer. Up to 5 samples/gel cassette may be run, with no possibility of cross contamination. Gel cassettes for DNA selections are available in 4 agarose concentrations to provide a full range of size selection options from 100bp up to 50kb. Run times range from ~30 minutes (small fragments) to several hours (pulsed-field runs for very large DNA fragments).
The BluePippin.jpg has the capabilities of the Pippin Prep, plus pulsed-field electrophoresis for resolving and collecting high molecular weight DNA. Target sizes or ranges of sizes are entered in software, and fractions are collected in buffer. Up to 5 samples/gel cassette may be run, with no possibility of cross contamination. Gel cassettes for DNA selections are available in 4 agarose concentrations to provide a full range of size selection options from 100bp up to 50kb. Run times range from ~30 minutes (small fragments) to several hours (pulsed-field runs for very large DNA fragments).
Cassette Kits
Gel Cassette Types and Target Ranges
Downstream Technologies
↵ Back to List of Instruments & selected Services
Client PC Workstation
 Advance Reservations, made through the Biosci-Booked (Genomics), are required for use of this computational resource (Computer-Workstation.jpg) in the Genomics Core. The computer has a variety of genomics-related software (see List) with associated User Guides, as well as standard Microsoft Office software. For descriptions of some of the ABI software, please see Data Analysis on the 'Services' page for the ABI 3130xl platform. Suggestions for additional 'genomics' software are welcome.
Advance Reservations, made through the Biosci-Booked (Genomics), are required for use of this computational resource (Computer-Workstation.jpg) in the Genomics Core. The computer has a variety of genomics-related software (see List) with associated User Guides, as well as standard Microsoft Office software. For descriptions of some of the ABI software, please see Data Analysis on the 'Services' page for the ABI 3130xl platform. Suggestions for additional 'genomics' software are welcome.
The Client PC Workstation is on LSU's network, and remote access to this resource will be granted on a case-by-case basis. Clients must adhere strictly to the schedule in Biosci-Booked (Genomics); otherwise, analyses might be lost when they log in while someone else is already using the computer. When accessing this resource from off-campus, your computer must have the LSU-approved VPN (gp.vpn.lsu.edu) installed and connected. Finally, clients must log on to the computer with their LSU credentials; however, such logins do not count against their allotment of "5 LSU PC registrations".
↵ Back to List of Instruments & selected Services
Accuris Series Tx (analytical balance)

↵ Back to List of Instruments & selected Services
QuantStudio·6 (qPCR)

Advance Reservations, made through the Biosci-Booked (Genomics) instrument reservation system, are required for use of this instrument. For a complete guide to qPCR experiments, see ThermoFisher's Real-time PCR Handbook 2.0 (2024; periodically updated). Our QuantStudio·6 has a standard 96-well block and is set up to use plates (not strip-tubes); thus, 0.2-ml 96-well plates are required. Calibrations on the machine were done with ABI plates (Thermofisher N801-0560) and might not be valid for plates from other vendors.
– Warning: v1.7 is
↵ Back to List of Instruments & selected Services
ABI Veriti (standard PCR thermal cycler)
 Advance Reservations, made through the Biosci-Booked (Genomics) instrument reservation system, are required for use of the ABI Veriti thermal cycler.jpg. This instrument performs standard PCR, including temperature gradients with 6 individualized sub-blocks, and it is protected from brief power outages by an APC backup power supply. Normally, all other standard thermocyclers in the Genomics Core are reserved solely for use by the Core or members of the Batzer lab; however, you may request permission to use the other standard thermocyclers if you can plead extenuating circumstances.
Advance Reservations, made through the Biosci-Booked (Genomics) instrument reservation system, are required for use of the ABI Veriti thermal cycler.jpg. This instrument performs standard PCR, including temperature gradients with 6 individualized sub-blocks, and it is protected from brief power outages by an APC backup power supply. Normally, all other standard thermocyclers in the Genomics Core are reserved solely for use by the Core or members of the Batzer lab; however, you may request permission to use the other standard thermocyclers if you can plead extenuating circumstances.
↵ Back to List of Instruments & selected Services
Qubit 4.0

The Qubit-4.0 (jpg) can quantify DNA, RNA, & proteins. In addition, with the Qubit RNA IQ assay, the Qubit 4.0 distinguishes intact from degraded RNA by using two unique dyes: one binds to large, intact RNA; the other binds to small, degraded RNA.
There are many Qubit assays available from ThermoFisher. Assays require 1-20 ul of sample, resulting in the quantitation ranges shown with each kit. Samples are analyzed using Axygen 0.5-ml tubes (PCR-05-C, thin-wall, clear).
If using your own kits, this instrument is available for use directly by you through the Biosci-Booked (Genomics) instrument reservation system; otherwise, samples are processed by the Genomics Core.
↵ Back to List of Instruments & selected Services
Diagenode Bioruptor NGS
 Advance Reservations, made through the Biosci-Booked (Genomics)
instrument reservation system, are required for use of this instrument. Further, prior to each use of this 'Reservation-Required' instrument, clients must ask the Genomics Core staff to prepare the system for operation.
Advance Reservations, made through the Biosci-Booked (Genomics)
instrument reservation system, are required for use of this instrument. Further, prior to each use of this 'Reservation-Required' instrument, clients must ask the Genomics Core staff to prepare the system for operation.
To retain the integrity of DNA, Diagenode’s Bioruptor® NGS uses a gentle method of sonication and a chilled water bath to generate indirect sonication waves. As a result, the Bioruptor® produces better and more consistent results than are achieved with harsher sonication methods. Up to 12 closed tubes can be sonicated in parallel and the continuous rotation of tubes allows even distribution of the energy for efficient sonication.
↵ Back to List of Instruments & selected Services
SpeedVac SPD1010
 Advance Reservations, made through the Biosci-Booked (Genomics)
instrument reservation system, are required for use of this instrument. This ThermoFisher instrument (SPD1010 SpeedVac.jpg) efficiently processes large batches of DNA or RNA samples. Currently, we have rotors for microtubes (1.5-ml, or 0.2-ml with inserts) and for 96-well plates. Typical applications include concentrating DNA extractions and removing low concentrations of non-agressive organic solvents (methanol, ethanol, acetonitrile) from samples. Most samples can be processed by simply pressing the "Start" button; however, users may also adjust settings for vacuum pressure, temperature, and run time.
Advance Reservations, made through the Biosci-Booked (Genomics)
instrument reservation system, are required for use of this instrument. This ThermoFisher instrument (SPD1010 SpeedVac.jpg) efficiently processes large batches of DNA or RNA samples. Currently, we have rotors for microtubes (1.5-ml, or 0.2-ml with inserts) and for 96-well plates. Typical applications include concentrating DNA extractions and removing low concentrations of non-agressive organic solvents (methanol, ethanol, acetonitrile) from samples. Most samples can be processed by simply pressing the "Start" button; however, users may also adjust settings for vacuum pressure, temperature, and run time.
↵ Back to List of Instruments & selected Services
Typhoon 8600 Variable Mode Imager
 Advance Reservations, made through the Biosci-Booked (Genomics) instrument reservation system, are required for use of this instrument. The Typhoon™ 8600 Variable Mode Imager is primarily used for storage phosphor experiments, but it is also capable of fluorescence detection of gels and blots. The Typhoon can scan storage phosphor screens (mounted or unmounted), gels and blots (≤ 35 × 43 cm). Images can be manipulated with ImageQuant™ Image Analysis Software; alternatively, users can change the file extension (i.e., *.gel) to "*.tif" and import the image into other software such as Adobe Photoshop.
Advance Reservations, made through the Biosci-Booked (Genomics) instrument reservation system, are required for use of this instrument. The Typhoon™ 8600 Variable Mode Imager is primarily used for storage phosphor experiments, but it is also capable of fluorescence detection of gels and blots. The Typhoon can scan storage phosphor screens (mounted or unmounted), gels and blots (≤ 35 × 43 cm). Images can be manipulated with ImageQuant™ Image Analysis Software; alternatively, users can change the file extension (i.e., *.gel) to "*.tif" and import the image into other software such as Adobe Photoshop.
↵ Back to List of Instruments & selected Services
E-gel (DNA Size-selection)
 The E-Gel® SizeSelect™ Gels are a convenient method for the purification of DNA libraries for Next
Generation Sequencing applications. DNA is size-selected in three easy steps: Load; Run; and, Retrieve.
The E-Gel® SizeSelect gel features two rows of wells. To extract a DNA region of interest without conventional
gel purification, simply load samples into the top row and electrophorese until the band (or smear of the
desired size range) moves into the bottom row of wells. Then, simply use a pipette to remove your purified
DNA (for smears, fragment sizes will have an ~50-bp range).
The E-Gel® SizeSelect™ Gels are a convenient method for the purification of DNA libraries for Next
Generation Sequencing applications. DNA is size-selected in three easy steps: Load; Run; and, Retrieve.
The E-Gel® SizeSelect gel features two rows of wells. To extract a DNA region of interest without conventional
gel purification, simply load samples into the top row and electrophorese until the band (or smear of the
desired size range) moves into the bottom row of wells. Then, simply use a pipette to remove your purified
DNA (for smears, fragment sizes will have an ~50-bp range).
Use of the E-gel can be either self-service (with permission) or staff-run. The Genomics Core usually stocks the 2% E-Gel®
SizeSelect™ Gels; however, other E-gel product options include:
- E-Gel EX agarose gels
- E-Gel NGS agarose gels
- E-Gel Clonewell agarose gels
- E-Gel 48/96 sample agarose gels
↵ Back to List of Instruments & selected Services
Eppendorf Centrifuge 5810R
 At this time, we do not require reservations for this instrument. The Eppendorf 5810R Centrifuge is a refrigerated unit (-9 to 40°C), capable of handling both large tubes and microtiter plates. Maximum speed is ~2800 rcf, and a soft acceleration-deceleration option is available. The Genomics Core has the A-4-62 rotor, which sports two types of adaptors: (A) microtiter plates or 96-well PCR plates; and, (B) 15-ml or 50-ml tubes (with correct inserts). Other rotors could be obtained if there is sufficient interest.
At this time, we do not require reservations for this instrument. The Eppendorf 5810R Centrifuge is a refrigerated unit (-9 to 40°C), capable of handling both large tubes and microtiter plates. Maximum speed is ~2800 rcf, and a soft acceleration-deceleration option is available. The Genomics Core has the A-4-62 rotor, which sports two types of adaptors: (A) microtiter plates or 96-well PCR plates; and, (B) 15-ml or 50-ml tubes (with correct inserts). Other rotors could be obtained if there is sufficient interest.
↵ Back to List of Instruments & selected Services
DNA Column Purification & Size Selection
 The Genomics Core stocks DNA columns by Zymo Research. Predominantly, we use two particular kits for 'column' Services, but Zymo kits designed for other applications can be ordered. For product specifications of all available kits, visit either the distributor's website (Genesee Scientific) or the manufacturer's website (Zymo Research).
The Genomics Core stocks DNA columns by Zymo Research. Predominantly, we use two particular kits for 'column' Services, but Zymo kits designed for other applications can be ordered. For product specifications of all available kits, visit either the distributor's website (Genesee Scientific) or the manufacturer's website (Zymo Research).
DNA Clean & Concentrator-5 Kit — (Cat #: 11-303):
Select-A-Size DNA Clean & Concentrator Kit — (Cat #D4080):
Other Zymo Kits (currently carried by Genomics Core):
↵ Back to List of Instruments & selected Services
SmartCheck
 The SmartCheck provides a fast, practical way to verify that a pipette is dispensing accurately (±5% at 10 µl, 20 µl, 50 µl, 100 µl, 200 µl, 300 µl and 1000 µl). Simply dispense distilled water into the opening and the test volume will be automatically detected. Repeat this three times to obtain a Pass/Fail result.
The SmartCheck provides a fast, practical way to verify that a pipette is dispensing accurately (±5% at 10 µl, 20 µl, 50 µl, 100 µl, 200 µl, 300 µl and 1000 µl). Simply dispense distilled water into the opening and the test volume will be automatically detected. Repeat this three times to obtain a Pass/Fail result.
Thus, you can both quickly verify your pipette's performance immediately prior to 'volume-critical' experiments and know when you really need to have your pipettes re-calibrated. The SmartCheck can also be used to train (or re-train!) personnel in the proper operation of pipettes.
↵ Back to List of Instruments & selected Services
Ancillary Support
 For a partial listing of the various smaller equipment and other ancillary items used to provide services at the LSU Genomics Core, see Ancillary Support. In addition to direct support equipment, the Genomics Core uses battery-backup units to ensure that instruments, equipment, and computers are protected from variable line voltage, brown-outs and temporary power outages. Further, the Core has installed air filtration products to protect instrumentation from sub-micron particulates that flow through the air handling system of the Life Sciences Annex (see How to improve Air Quality in your lab?).
For a partial listing of the various smaller equipment and other ancillary items used to provide services at the LSU Genomics Core, see Ancillary Support. In addition to direct support equipment, the Genomics Core uses battery-backup units to ensure that instruments, equipment, and computers are protected from variable line voltage, brown-outs and temporary power outages. Further, the Core has installed air filtration products to protect instrumentation from sub-micron particulates that flow through the air handling system of the Life Sciences Annex (see How to improve Air Quality in your lab?).
↵ Back to List of Instruments & selected Services
Legacy instruments
 The instruments at Legacy instruments are physically still present in the Core's inventory. However, their usage is either not possible or significant preparatory work & expense will be required to bring them online.
The instruments at Legacy instruments are physically still present in the Core's inventory. However, their usage is either not possible or significant preparatory work & expense will be required to bring them online.
Thus, if you are interested in those instruments, please contact the Genomics Core for further information. Alternatively, if you wish to propose that a new instrument be purchased for the Core, follow the instructions at Potential Instrument Acquisitions.

